DICOM Query Retrieve Service¶
To query retrieve dose related objects from a remote server, you need to review the DICOM Network Configuration first.
Query-retrieve using the web interface¶
- On the Imports menu, select
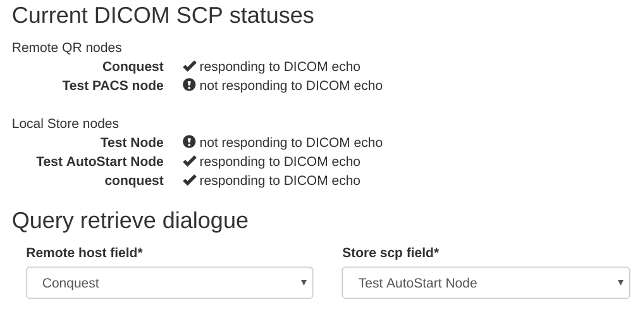
Query remote server- see figure 1. If the menu isn’t there, you need to check your user permissions - see Configure the settings for details. - Each configured query-retrieve node and each local store node is automatically tested to make sure they respond to a DICOM echo - the results are presented at the top of the page. See fingure 2 for an example.
- Select the desired remote host, ie the PACS or modality you wish to query.
- Select the local store node you want to retrieve to.
- Select which modalities you want to query for - at least one must be ticked.
- Select a date range - the wider this is, the more stress the query will place on the remote server, and the higher the likelyhood of the query being returned with zero results (a common configuration on the remote host to prevent large database queries affecting other services).
- If you wish to exclude studies based on their study description, enter the text here. Add several terms by separating
them with a comma. One example would be to exclude any studies with
importedin the study description, if your institution modifies this field on import. The matching is case-insensitive. - Alternatively, you might want to only keep studies with particular terms in the study description. If so, enter them in the next box, comma separated.
Advanced query options¶
- Include SR only studies default not ticked: If you have a DICOM store with only the radiation dose structured reports (RDSR) in, or a mix of whole studies and RDSRs without the corresponding study, then tick this box.
- Ignore studies already in the database default ticked: The RDSR import routine checks for the existance of the study UID in the database, and if it is found they it doesn’t go any further. This might change in the future as there are instances where two RDSRs might legitimately have the same study UID, but different event UIDs. For image based imports, the individual events are checked, so if you think there is a reasonable chance that the database is missing individual images from a study, then you might like to deselect this setting. If the same dates are selected multiple times (to update during a day for example), activating this setting will result in the same exams all being transferred each time.
When you have finished the query parameters, click Submit
Review and retrieve¶
The progress of the query is reported on the right hand side. If nothing happens, ask the adminsitrator to check if the celery queue is running.
Once all the responses have been purged of unwanted modalities, study descriptions or study UIDs, the number of studies
of each type will be displayed and a button appears. Click Retreive to request the remote server send the selected
objects to your selected Store node. This will be based on your original selection - changing the node on the left hand
side at this stage will have no effect.
The progress of the retrieve is displayed in the same place until the retrieve is complete.
Query-retrieve using the command line interface¶
In a command window/shell, qrscu.py -h should present you with the following output:
usage: qrscu.py [-h] [-ct] [-mg] [-fl] [-dx] [-f yyyy-mm-dd] [-t yyyy-mm-dd]
[-e string] [-i string] [-sr] [-dup] qrid storeid
Query remote server and retrieve to OpenREM
positional arguments:
qrid Database ID of the remote QR node
storeid Database ID of the local store node
optional arguments:
-h, --help show this help message and exit
-ct Query for CT studies
-mg Query for mammography studies
-fl Query for fluoroscopy studies
-dx Query for planar X-ray studies
-f yyyy-mm-dd, --dfrom yyyy-mm-dd
Date from, format yyyy-mm-dd
-t yyyy-mm-dd, --duntil yyyy-mm-dd
Date until, format yyyy-mm-dd
-e string, --desc_exclude string
Terms to exclude in study description, comma separated, quote whole
string
-i string, --desc_include string
Terms that must be included in study description, comma separated,
quote whole string
-sr Advanced: Query for structured report only studies
-dup Advanced: Retrieve studies that are already in database
As an example, if you wanted to query the PACS for DX images on the 5th April 2010 with any study descriptions including
imported excluded, first you need to know the database IDs of the remote node and the local node you want the images
sent to. To find these, go to the DICOM Network Configuration page where the database ID is listed among the other details for
each node.
Assuming the PACS database ID is 2, and the store node ID is 1, the command would look something like:
qrscu.py 2 1 -dx -f 2010-04-05 -t 2010-04-05 -e "imported"
If you want to do this regularly to catch new studies, you might like to use a script something like this on linux:
#!/bin/bash
. /var/openrem/bin/activate # activate virtualenv if you are using one, modify or delete this line
ONEHOURAGO=$(date -d "1 hour ago" "+%Y-%m-%d")
openrem_qr.py 2 1 -dx -f $ONEHOURAGO -t $ONEHOURAGO -e "Imported"
This script could be run once an hour using a cron job. By asking for the date an hour ago, you shouldn’t miss exams taking place in the last hour of the day.
A similar script could be created as a batch file on Windows and run using the scheduler.
Troubleshooting: openrem_qr.log¶
If the default logging settings haven’t been changed then there will be a log files to refer to. The default
location is within your MEDIAROOT folder:
This file contains information about the query, the status of the remote node, the C-Find response, the analysis of the response, and the individual C-Move requests.
The following is an example of the start of the log for the following query which is run once an hour (ie some responses will already have been imported):
qrscu.py 2 1 -dx -f 2016-05-04 -t 2016-05-04 -e "imported"
[04/May/2016 11:30:02] INFO [remapp.netdicom.qrscu:580] qrscu script called
[04/May/2016 11:30:02] INFO [remapp.netdicom.qrscu:595] Modalities are ['DX']
[04/May/2016 11:30:02] INFO [remapp.netdicom.qrscu:601] Date from: 2016-05-04
[04/May/2016 11:30:02] INFO [remapp.netdicom.qrscu:604] Date until: 2016-05-04
[04/May/2016 11:30:02] INFO [remapp.netdicom.qrscu:610] Study description exclude terms are ['imported']
[04/May/2016 11:30:03] INFO [remapp.netdicom.qrscu:267] Request association with Hospital PACS PACSAET01 (PACSEAT01 104 DICOM_QR_SCP)
[04/May/2016 11:30:03] INFO [remapp.netdicom.qrscu:33] Association response received
[04/May/2016 11:30:03] INFO [remapp.netdicom.qrscu:277] assoc is ... <Association(Thread-7208, started daemon 140538998306560)>
[04/May/2016 11:30:03] INFO [remapp.netdicom.qrscu:280] DICOM Echo ...
[04/May/2016 11:30:03] INFO [remapp.netdicom.qrscu:282] done with status Success
[04/May/2016 11:30:03] INFO [remapp.netdicom.qrscu:284] DICOM FindSCU ...
[04/May/2016 11:30:03] INFO [remapp.netdicom.qrscu:311] Currently querying for DX studies...
[04/May/2016 11:30:03] INFO [remapp.netdicom.qrscu:33] Association response received
[04/May/2016 11:30:04] INFO [remapp.netdicom.qrscu:33] Association response received
[04/May/2016 11:30:04] INFO [remapp.netdicom.qrscu:33] Association response received
[04/May/2016 11:30:04] INFO [remapp.netdicom.qrscu:33] Association response received
[04/May/2016 11:30:05] INFO [remapp.netdicom.qrscu:33] Association response received
[04/May/2016 11:30:05] INFO [remapp.netdicom.qrscu:311] Currently querying for CR studies...
[04/May/2016 11:30:05] INFO [remapp.netdicom.qrscu:33] Association response received
[04/May/2016 11:30:05] INFO [remapp.netdicom.qrscu:33] Association response received
[04/May/2016 11:30:06] INFO [remapp.netdicom.qrscu:33] Association response received
[04/May/2016 11:30:06] INFO [remapp.netdicom.qrscu:33] Association response received
[04/May/2016 11:30:06] INFO [remapp.netdicom.qrscu:33] Association response received
[04/May/2016 11:30:07] INFO [remapp.netdicom.qrscu:33] Association response received
[04/May/2016 11:30:10] INFO [remapp.netdicom.qrscu:33] Association response received
[04/May/2016 11:30:10] INFO [remapp.netdicom.qrscu:33] Association response received
[04/May/2016 11:30:11] INFO [remapp.netdicom.qrscu:33] Association response received
[04/May/2016 11:30:11] INFO [remapp.netdicom.qrscu:33] Association response received
[04/May/2016 11:30:12] INFO [remapp.netdicom.qrscu:33] Association response received
[04/May/2016 11:30:12] INFO [remapp.netdicom.qrscu:33] Association response received
[04/May/2016 11:30:12] INFO [remapp.netdicom.qrscu:33] Association response received
[04/May/2016 11:30:12] INFO [remapp.netdicom.qrscu:339] Checking to see if any of the 16 studies are already in the OpenREM database
[04/May/2016 11:30:13] INFO [remapp.netdicom.qrscu:343] Now have 11 studies
[04/May/2016 11:30:13] INFO [remapp.netdicom.qrscu:349] Deleting studies we didn't ask for
[04/May/2016 11:30:13] INFO [remapp.netdicom.qrscu:358] mod is DX, mod_set is ["CR"]
[04/May/2016 11:30:13] INFO [remapp.netdicom.qrscu:358] mod is CR, mod_set is ["CR"]
[04/May/2016 11:30:13] INFO [remapp.netdicom.qrscu:358] mod is DX, mod_set is ["PR", "DX"]
[04/May/2016 11:30:13] INFO [remapp.netdicom.qrscu:358] mod is CR, mod_set is ["PR", "DX"]
[04/May/2016 11:30:13] INFO [remapp.netdicom.qrscu:358] mod is DX, mod_set is ["DX"]
[04/May/2016 11:30:13] INFO [remapp.netdicom.qrscu:358] mod is CR, mod_set is ["DX"]
[04/May/2016 11:30:13] INFO [remapp.netdicom.qrscu:358] mod is DX, mod_set is ["PR", "CR"]
[04/May/2016 11:30:13] INFO [remapp.netdicom.qrscu:358] mod is CR, mod_set is ["PR", "CR"]
[04/May/2016 11:30:13] INFO [remapp.netdicom.qrscu:367] Now have 11 studies
[04/May/2016 11:30:13] INFO [remapp.netdicom.qrscu:372] Deleting series we can't use
[04/May/2016 11:30:13] INFO [remapp.netdicom.qrscu:408] Now have 11 studies
[04/May/2016 11:30:13] INFO [remapp.netdicom.qrscu:413] Deleting any studies that match the exclude criteria
[04/May/2016 11:30:13] INFO [remapp.netdicom.qrscu:422] Now have 6 studies after deleting any containing any of [u'imported']
[04/May/2016 11:30:13] INFO [remapp.netdicom.qrscu:438] Release association
[04/May/2016 11:30:13] INFO [remapp.netdicom.qrscu:499] Preparing to start move request
[04/May/2016 11:30:13] INFO [remapp.netdicom.qrscu:504] Requesting move of 6 studies
[04/May/2016 11:30:13] INFO [remapp.netdicom.qrscu:509] Mv: study_no 1
[04/May/2016 11:30:13] INFO [remapp.netdicom.qrscu:515] Mv: study no 1 series no 1
[04/May/2016 11:30:13] INFO [remapp.netdicom.qrscu:528] Requesting move: modality DX, study 1 (of 6) series 1 (of 1). Series contains 1 objects
[04/May/2016 11:30:13] INFO [remapp.netdicom.qrscu:33] Association response received
[04/May/2016 11:30:13] INFO [remapp.netdicom.qrscu:44] Move association requested
[04/May/2016 11:30:18] INFO [remapp.netdicom.qrscu:53] Move association released
[04/May/2016 11:30:18] INFO [remapp.netdicom.qrscu:532] _move_req launched
[04/May/2016 11:30:18] INFO [remapp.netdicom.qrscu:509] Mv: study_no 2
[04/May/2016 11:30:18] INFO [remapp.netdicom.qrscu:515] Mv: study no 2 series no 1
[04/May/2016 11:30:18] INFO [remapp.netdicom.qrscu:528] Requesting move: modality DX, study 2 (of 6) series 1 (of 1). Series contains 2 objects
[04/May/2016 11:30:18] INFO [remapp.netdicom.qrscu:33] Association response received
[04/May/2016 11:30:19] INFO [remapp.netdicom.qrscu:44] Move association requested
[04/May/2016 11:30:29] INFO [remapp.netdicom.qrscu:48] gg is Pending
[04/May/2016 11:30:30] INFO [remapp.netdicom.qrscu:53] Move association released
...etc
If you are using an OpenREM native storage node, then you might also like to review Troubleshooting: openrem_store.log